December 2022
Decipher Bladder Genomic Classifier
CLINICAL CANCER RESEARCH
Neoadjuvant pembrolizumab and radical cystectomy in patients with muscle-invasive urothelial bladder cancer: 3-year median follow-up update of PURE-01 trial
Basile G, et alDECIPHER® PROSTATE METASTATIC TEST NOW AVAILABLE
Expanded access for patients with metastatic prostate cancer in the U.S.
Our goal is to help physicians and patients feel well-informed throughout the cancer care journey, by sharing new insights and more information about Veracyte’s urologic cancer genomic tests, Decipher Prostate and Decipher Bladder.


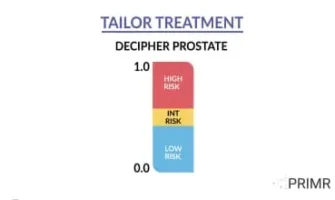

Download the test report samples for the Decipher Prostate and Decipher Bladder Genomic Classifiers
Go to Test Report Page
Download the requisition and shipping Instructions for the Decipher Prostate and Decipher Bladder Genomic Classifiers
Go to Requisitions Page
Download the patient and physician brochures for the Decipher Prostate Genomic Classifier in English and Spanish
Go to Brochures Page
View our Local Coverage Determination for Decipher Prostate
Go to LCD for Decipher Prostate
View our Local Coverage Determination for Decipher Bladder
Go to LCD for Decipher Bladder

We are dedicated to the continued pursuit of evidence. The Decipher Prostate Genomic Classifier has been validated in over 70 studies of more than 100,000 patients as a highly accurate predictor of metastasis, prostate cancer-specific mortality, and overall survival.
December 2022
Decipher Bladder Genomic Classifier
CLINICAL CANCER RESEARCH
Neoadjuvant pembrolizumab and radical cystectomy in patients with muscle-invasive urothelial bladder cancer: 3-year median follow-up update of PURE-01 trial
Basile G, et alMarch 2023
Decipher Prostate RP
THE BRITISH JOURNAL OF RADIOLOGY
The combination of prostate MRI PI-RADS scoring system and a genomic classifier is associated with pelvic lymph node metastasis at the time of radical prostatectomy
Benedir T, et alApril 2023
Decipher Prostate Biopsy
European Urology Oncology
Transcriptomic Heterogeneity in High-risk Prostate Cancer and Implications for Extraprostatic Disease at Presentation on Prostate-specific Membrane Antigen Positron Emission Tomography
Smith CP, et alJuly 2023
Reviews including Decipher
THE INTERNATIONAL JOURNAL OF RADIATION
Genomic classifiers in personalized prostate cancer radiotherapy approaches – a systematic review and future perspectives based on international consensus
Spohn S, et alAugust 2023
Decipher GRID
JCO PRECISION ONCOLOGY
Transcriptomic Signatures Associated With Outcomes in Recurrent Prostate Cancer Treated With Salvage Radiation, Androgen-Deprivation Therapy, and Enzalutamide: Correlative Analysis of the STREAM Trial
Bitting RL, et alReferences